library(tidyverse)
library(sf)
library(terra)
library(stars)
library(gstat)
library(tmap)
library(kableExtra)ca_ecoregions <- st_read(here::here("data", "CA_Level3_Ecoregions_EPA", "ca_eco_l3.shp"))
bioclim_dir <- here::here("data", "wc2.1_2.5m")
bioclim <- list.files(bioclim_dir, pattern = glob2rx("*.tif$"), full.names = TRUE)
bioclim_sort <- bioclim[
# Sort filepaths based on numeric suffix
order(
# Extract numeric suffix of filenames and convert to numeric
as.numeric(gsub(".*_(\\d+)\\.tif$", "\\1", bioclim)))]
bioclim_rast <- rast(bioclim_sort)
variables <- c("annualMeanTemp", "meanDiurnalRange", "isothermality", "tempSeasonality", "maxTempWarmMonth", "maxTempColdMonth", "tempAnnualRange", "meanTempWetQ", "meanTempDryQ", "meanTempWarmQ", "meanTempColdQ", "annualPrecip", "precipWetMonth", "precipDryMonth", "precipSeasonality", "precipWetQ", "precipDryQ", "precipWarmQ", "precipColdQ")
names(bioclim_rast) <- variablesTransform CRS
ca_ecoregions <- ca_ecoregions %>%
st_transform(crs = st_crs(bioclim_rast))Crop and Mask Raster
bioclim_rast_crop <- crop(bioclim_rast, ca_ecoregions)
bioclim_rast_mask <- mask(bioclim_rast_crop, ca_ecoregions)Sample Random Points in Raster
set.seed(275)
pts <- spatSample(bioclim_rast_mask,
# Random sample non-NA cells
na.rm = TRUE,
# Sample size
size = 1000,
# Return SpatVector
as.points = TRUE)
pts_sf <- st_as_sf(pts)tm_shape(bioclim_rast_mask[["annualMeanTemp"]]) +
tm_raster(palette = "-RdYlBu", title = "Annual Mean Temp") +
tm_shape(pts_sf) +
tm_dots(col = "#403d39", size = 0.075,
title = "Random Samples") +
tm_add_legend(type = "symbol", labels = "Random Samples",
col = "#403d39", size = 0.075) +
tm_layout(legend.position = c("right", "top"),
legend.bg.color = "white")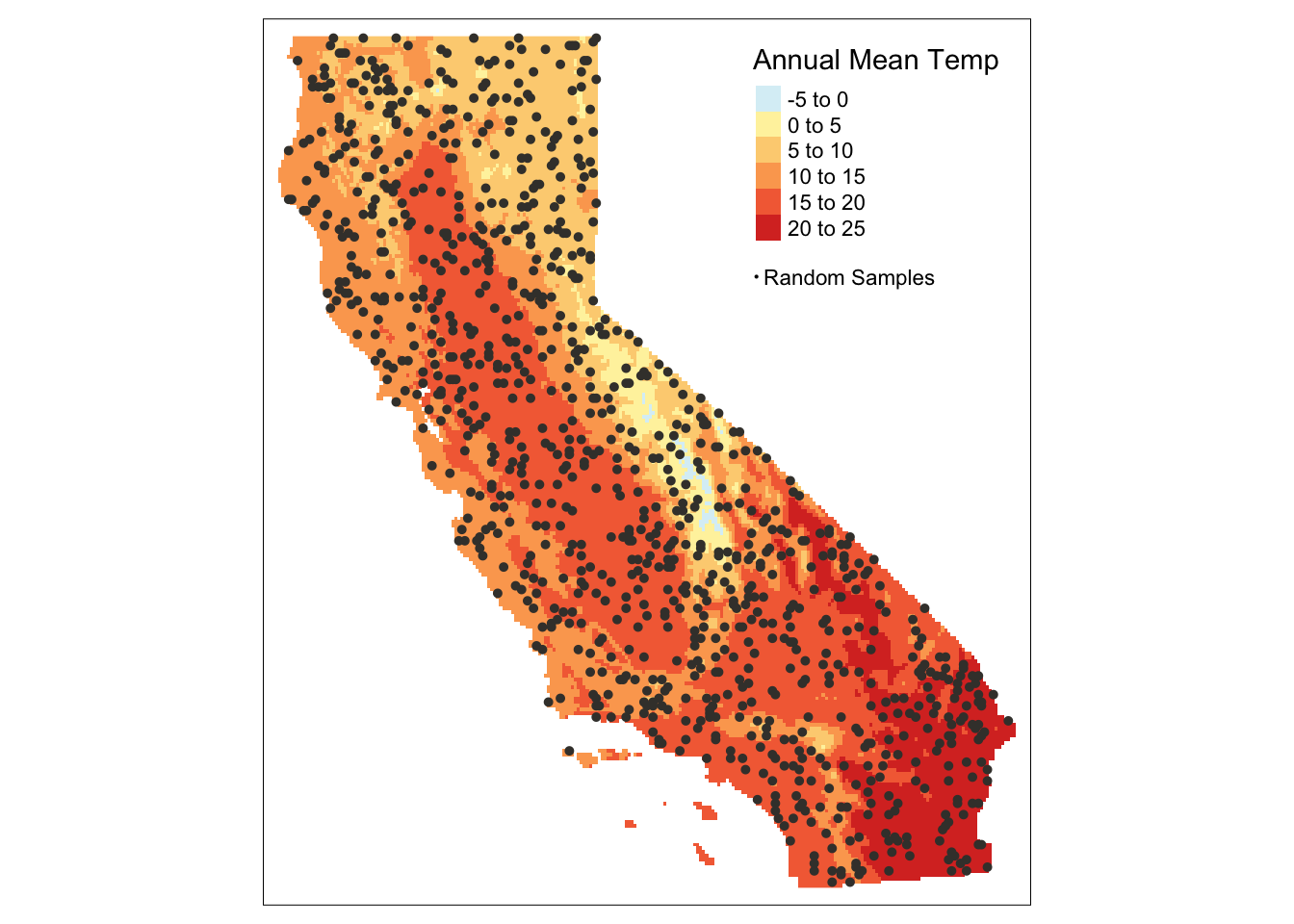
Perform IDW Interpolation
grid <- st_make_grid(pts_sf)
grid <- stars::st_as_stars(grid, crs = st_crs(pts_sf))annualMeanTemp_idw <- idw(annualMeanTemp ~ 1, locations = pts_sf, newdata = grid, idp = 2)View IDW Interpolation
annualMeanTemp_idw <- rast(annualMeanTemp_idw)
annualMeanTemp_idw <- mask(annualMeanTemp_idw, ca_ecoregions)map_1 <- tm_shape(bioclim_rast_mask[["annualMeanTemp"]]) +
tm_raster(palette = "-RdYlBu", title = "Annual Mean Temp") +
tm_layout(legend.position = c("right", "top"),
legend.bg.color = "white")
map_2 <- tm_shape(ca_ecoregions) +
tm_polygons() +
tm_shape(pts_sf) +
tm_dots(col = "#403d39", size = 0.075,
title = "Random Samples") +
tm_add_legend(type = "symbol", labels = "Random Samples",
col = "#403d39", size = 0.075) +
tm_layout(legend.position = c("right", "top"),
legend.bg.color = "white")
map_3 <- tm_shape(annualMeanTemp_idw[["var1.pred_var1.pred"]]) +
tm_raster(palette = "-RdYlBu", title = "Annual Mean Temp") +
tm_layout(legend.position = c("right", "top"),
legend.bg.color = "white")
tmap_arrange(map_1, map_2, map_3)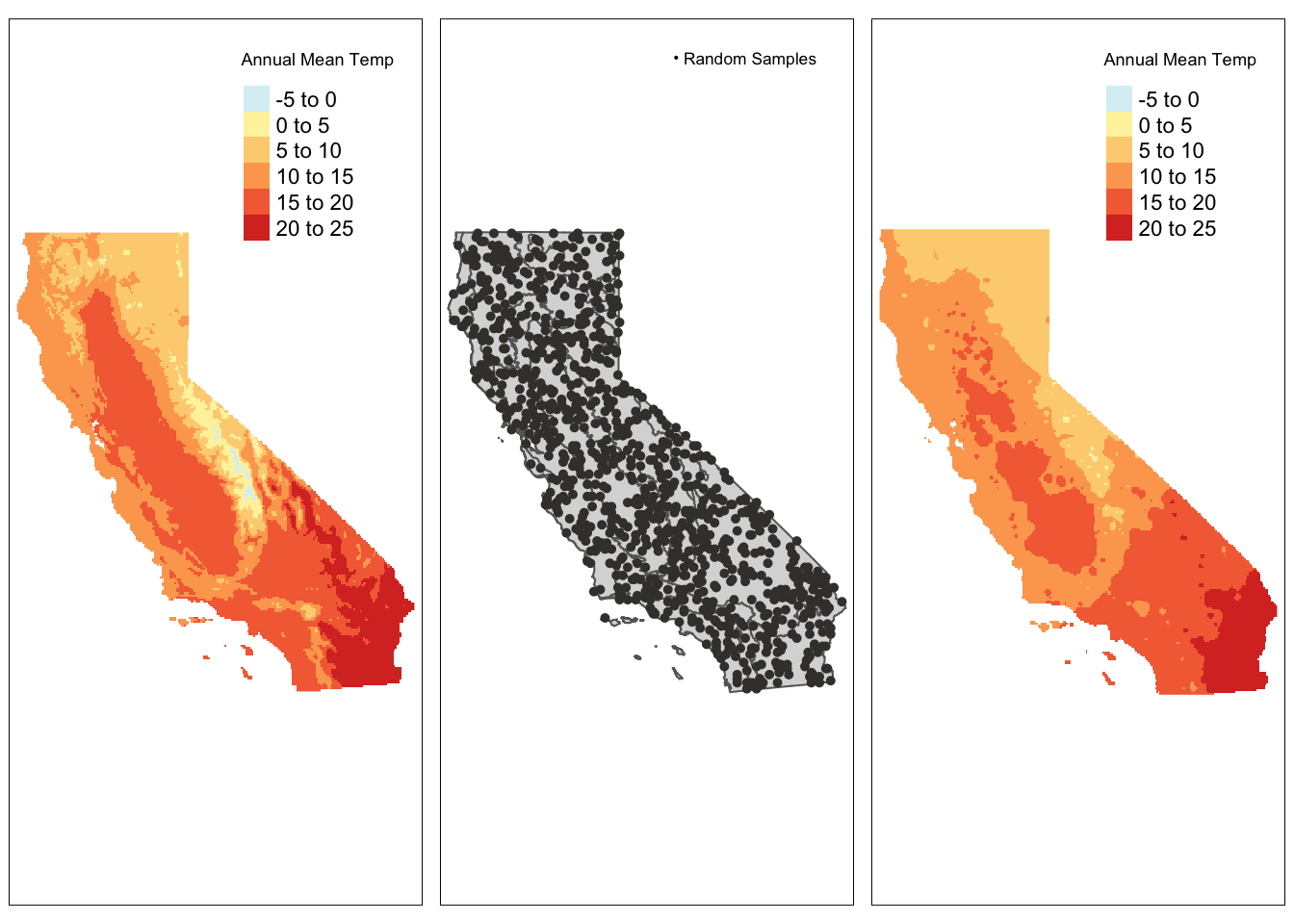
Find Interpolated Mean for Each Ecoregion
grid_rast <- rast(grid)
ca_ecoregions_rast <- rasterize(ca_ecoregions, grid_rast, field = "US_L3NAME")tm_shape(ca_ecoregions_rast) +
tm_raster(title = "Ecoregions") +
tm_layout(legend.outside = TRUE,
legend.title.size = 0.75,
legend.text.size = 0.75,
legend.outside.size = 0.55,
legend.bg.color = "white")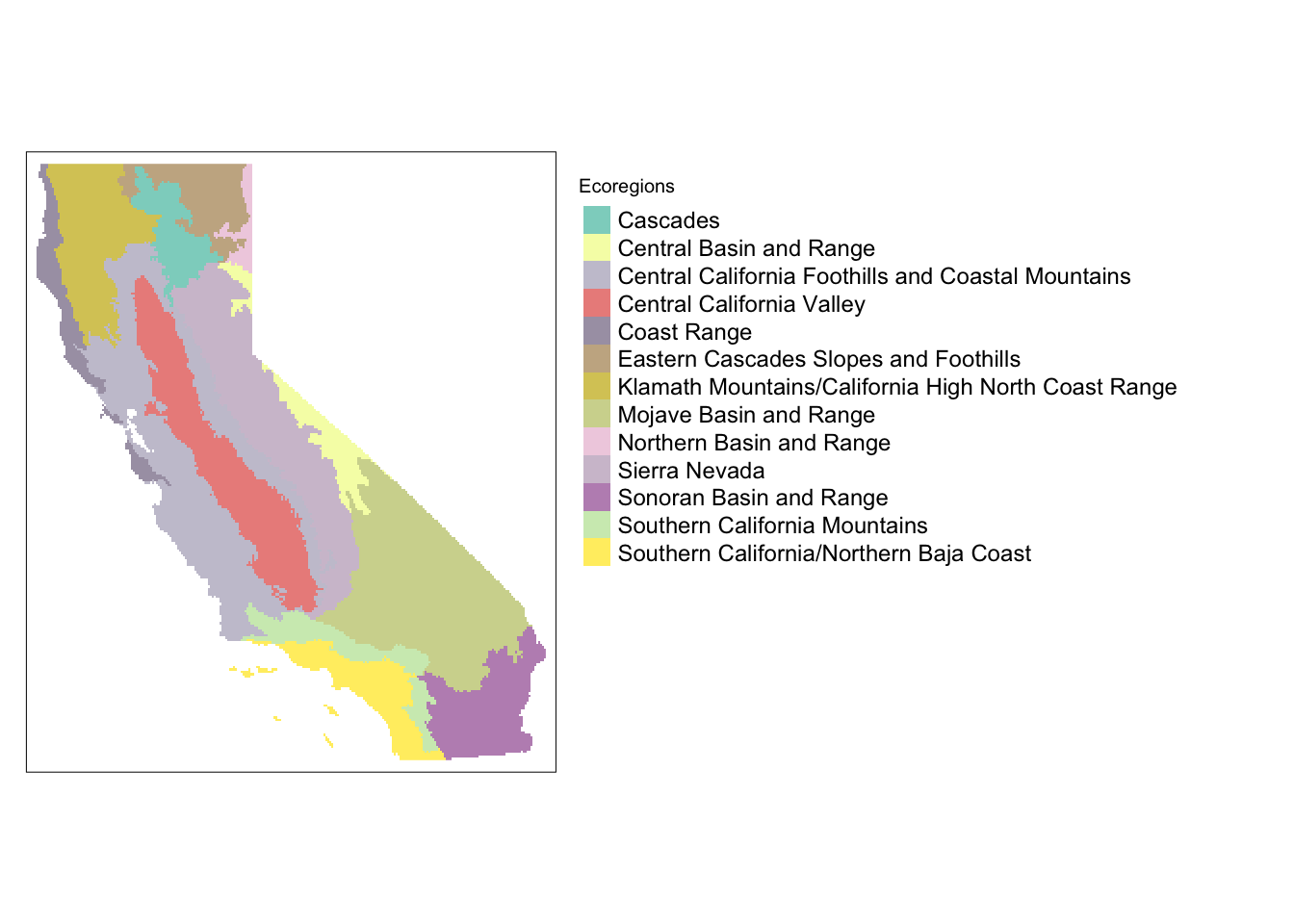
pred_by_ecoregion <- zonal(annualMeanTemp_idw, ca_ecoregions_rast, fun = "mean", na.rm = TRUE)pred_by_ecoregion %>%
rename(ecoregion = US_L3NAME,
mean_annualMeanTemp_pred = var1.pred_var1.pred) %>%
select(ecoregion, mean_annualMeanTemp_pred) %>%
kbl() %>%
kable_minimal()| ecoregion | mean_annualMeanTemp_pred |
|---|---|
| Cascades | 9.268085 |
| Central Basin and Range | 10.085137 |
| Central California Foothills and Coastal Mountains | 14.465699 |
| Central California Valley | 15.481970 |
| Coast Range | 12.190697 |
| Eastern Cascades Slopes and Foothills | 8.471113 |
| Klamath Mountains/California High North Coast Range | 10.413797 |
| Mojave Basin and Range | 17.363583 |
| Northern Basin and Range | 8.532363 |
| Sierra Nevada | 10.134800 |
| Sonoran Basin and Range | 21.047092 |
| Southern California Mountains | 15.140603 |
| Southern California/Northern Baja Coast | 16.610146 |